Research
The ultimate goal of our lab is to develop novel successful therapies for children with brain tumors. By dissecting the fundamental processes of tumor biology and by developing relevant mouse models we have powerful tools in hand to attain this goal. In our lab we focus our efforts on three key areas:
- Identify and validate the importance of mutations in pHGG
- Understand why these mutations drive tumor formation.
- Do these mutations confer a Therapeutic Vulnerability?
Pediatric High Grade Glioma (pHGG) is a devastating disease with a median survival of ~12 months. Despite a substantial number of clinical trials, patient outcome has barely improved. Crucially, pHGG are very different from their adult counterparts, not only in their mutational burden but also in their therapeutic response. To design successful clinical trials it is imperative we perform high quality pre-clinical studies in genetically accurate pHGG mouse models with an intact immune system. Currently however, these models do not exist, not only hampering our ability to preclinically evaluate potential therapies but also impeding our ability to mechanistically dissect how these pHGG form.
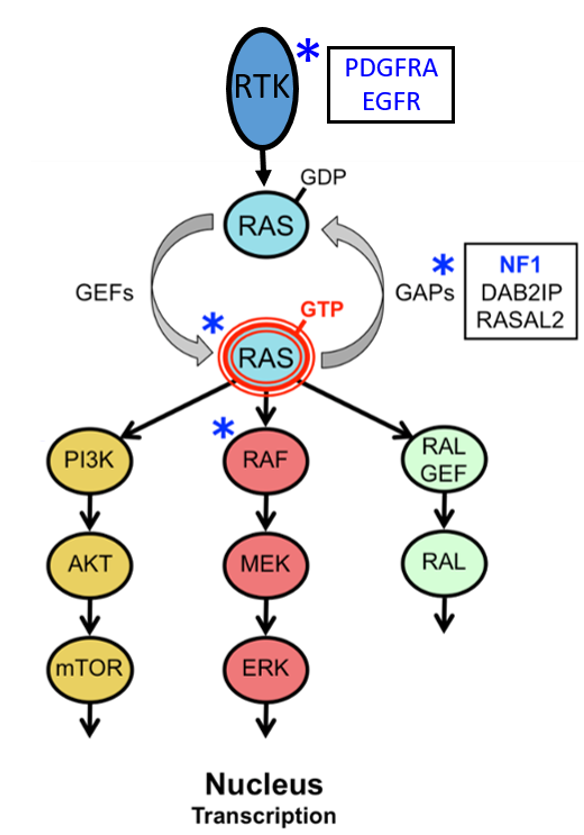
Intruigingly, these pHGG often have abberant activation of the RAS pathway. Given our interest in RAS pathway signaling and cancer, a subset of our research focuses on the RasGAP NF1. The NF1 gene is a tumor suppressor that negatively regulates RAS. NF1 is the causal gene of the common familial tumor predisposition syndrome Neurofibromatosis type I (NF1). NF1 patients develop an array of both benign and malignant central and peripheral nervous system tumors, all of which display aberrant RAS pathway activation. Of these, neurofibromas (benign), low-grade gliomas and Malignant Peripheral Nerve Sheath Tumors (MPNSTs) are the most prominent; for none of these a cure exists. Importantly, NF1 is also frequently mutated in sporadic cancers such as glioblastoma multiforme (GBM), pediatric high grade gliomas, melanoma and lung cancer.
Identify and Validate the importance of cooperating mutations in pHGG
In the current era of next generation sequencing, identifying potential new drivers for cancer is no longer a bottleneck. A major challenge, however, remains the rapid functional validation of the vast number of genes that were found mutated or lost. With regards to functional validation, in vivo modeling remains a state of the art discovery tool. However, the development of classical genetically engineered mouse models is cumbersome and time consuming, especially when several genetic drivers need to be combined.
We are using both traditional (stereotactic injection of modified neuronal stem cells) and more advanced (stereotactic injection of a CRISPR delivery system) in vivo modeling systems to screen cooperating candidate tumor suppressor and oncogenes that drive central nervous system tumors. A key element of both systems is that they are Fast, Flexible and Tractable. These systems thus allow us to generate more genetically accurate mouse models for central nervous system tumors.
We have a keen interest in investigating the cooperativity between loss of NF1 and mutations in the epigenetic machinery. Intriguingly, brain tumors often have mutations in epigenetic drivers.
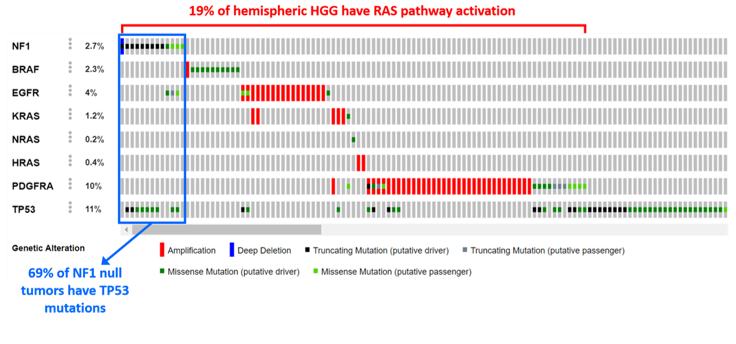
Understand why mutations drive tumor formation
Crucially, if we want to develop rational therapies for patients with pHGG we will have to study and understand the intricacies of how mutations in specific genes cooperate and discover which pathways are crucial for progression or therapeutic intervention. This focus on a basic research component is essential for success.
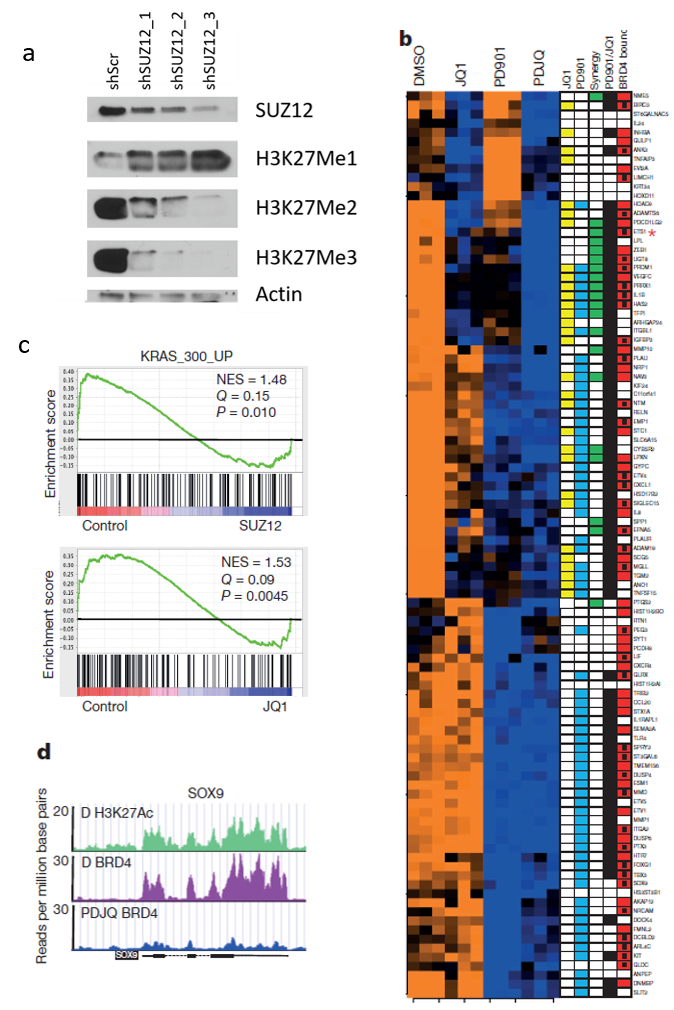
Our lab uses a multitude of techniques, ranging from RNAseq to ChIPseq, CRISPR and mouse modeling, to answer the “why” and “how” questions that are so crucial to enhance our understanding of what drives these pediatric brain tumors. Essential for these endeavors are research samples. Importantly, Children’s Hospital Philadelphia is the home of the CBTTC, the Children’s Brain Tumor Tissue Consortium, a consortium that collects, sequences and distributes samples and data of pHGG. Through the CBTTC we already have access to a wealth of the tumor samples needed for our research.
We have a keen interest in investigating the cooperativity between loss of NF1 and mutations in the epigenetic machinery. A large portion of our work hence focuses on the question of how exactly these epigenetic drivers promote tumor formation in pHGG with NF1 loss.
Preclinical testing of novel therapies
If we want to develop and test effective therapies, there is a need to generate mouse models that can keep up with our continuously evolving understanding of the genetic and epigenetic events that drive cancer. Moreover, it is imperative that we not only generate genetically accurate mouse models for these gliomas but also phenocopy the tumor microenvironment (i.e. model pHGG in mice with an intact immune system) as the tumor microenvironment undoubtedly plays an important role during the formation of these tumors. Our mouse models will not only allow us to study the biology in vivo but will also allow us to test therapies (including immunotherapies) that, when successful, could be translated into clinical trials. Notably, we have over a decade of expertise in mouse modeling and pre-clinical trial development (De Raedt et al. Nature 2014) and our work has been translated into several clinical trials.
Immunotherapy and Immune Micro-environment
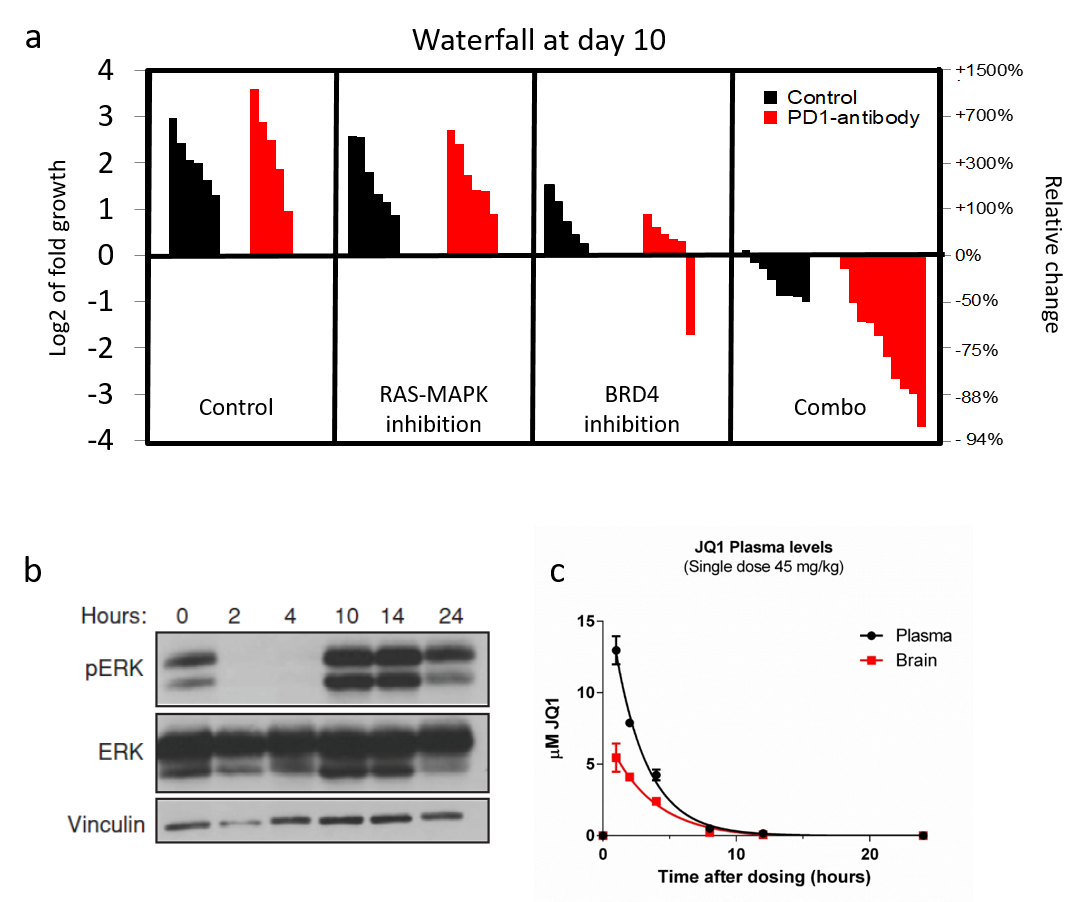
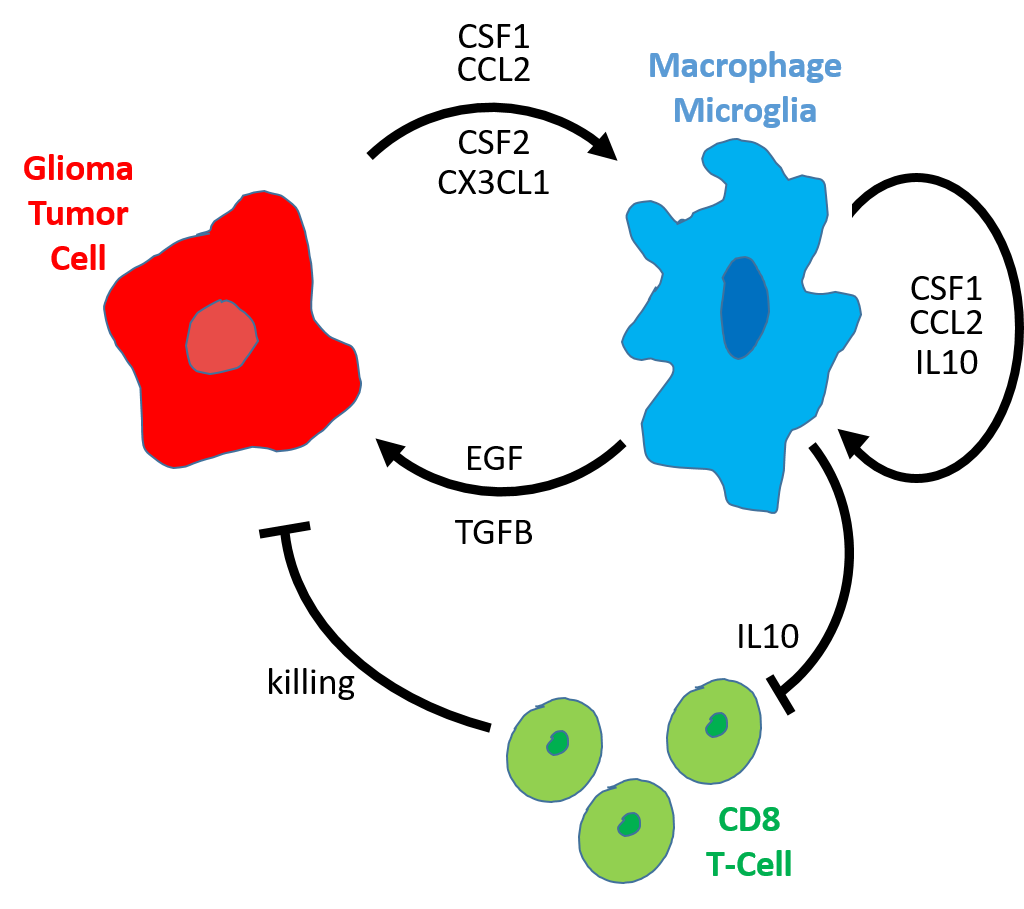
Infiltrating immune cells play an important role during glioma formation. Some of these immune cells have an anti-tumor function, importantly however, others actually protect and stimulate the glioma tumor cells. Crucially, we have developed mouse model of pHGG that accurately models human pediatric High Grade Gliomas and have both the infiltrating “good” (CD8 T-cells) and “bad” (microglia) immune cells.We will test a combination therapy in these mice that, in addition to targeting the tumor cells directly, stimulates the CD8 T-immune cells while inhibiting the pro-tumor microglia immune cells. If our approach is successful this therapy can quickly be translated into the clinic as all drugs used are approved or being used in clinical trials.
An important part of our research focuses on the immune microenvironment. We will test how important certain infiltrating immune (“good” or “bad”) are for pediatric glioma formation. We will not only determine if we can target these immune cells and cause tumors to shrink, but we will also able to dissect the intricacies of how exactly these immune cells behave in the tumor. We will answer questions like: How exactly do the “bad” microglia stimulate tumor formation? And how exactly do the tumor cells recruit and manipulate the immune system for their advantage?
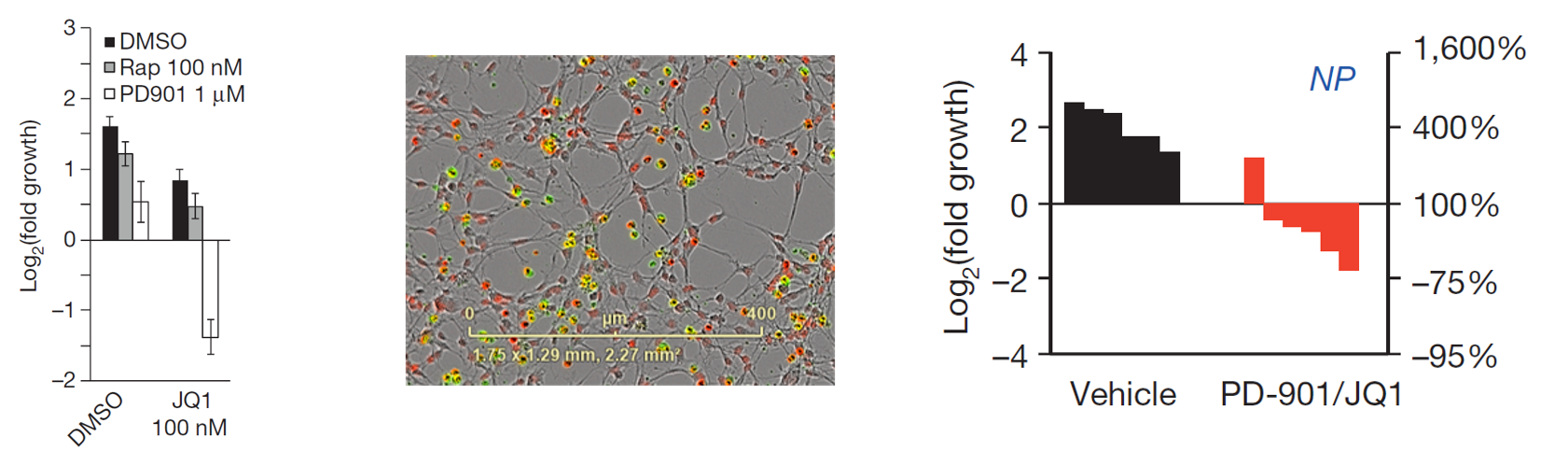
Epigenetic Based Therapies
We have a keen interest in investigating the cooperativity between loss of NF1 and mutations in the epigenetic machinery. These epigenetic mutations often make tumors hyper sensitive to epigenetic based therapies. We have had a lot of success by using MEK (PD-0325901) in combination with BRD4 inhibitors (JQ1).
Select Publication
-
2017
Olsen Sarah Naomi, Wronski Ania, Castaño Zafira, Dake Benjamin, Malone Clare, De Raedt Thomas, Enos Miriam, DeRose Yoko S, Zhou Wenhui, Guerra Stephanie, Loda Massimo, Welm Alana, Partridge Ann H, McAllister Sandra S, Kuperwasser Charlotte, Cichowski Karen: Loss of RasGAP Tumor Suppressors Underlies the Aggressive Nature of Luminal B Breast Cancers. Cancer discovery 7(2): 202-217, Feb 2017.
2016
Zhang Haikuo, Qi Jun, Reyes Jaime M, Li Lewyn, Rao Prakash K, Li Fugen, Lin Charles Y, Perry Jennifer A, Lawlor Matthew A, Federation Alexander, De Raedt Thomas, Li Yvonne Y, Liu Yan, Duarte Melissa A, Zhang Yanxi, Herter-Sprie Grit S, Kikuchi Eiki, Carretero Julian, Perou Charles M, Reibel Jacob B, Paulk Joshiawa, Bronson Roderick T, Watanabe Hideo, Brainson Christine Fillmore, Kim Carla F, Hammerman Peter S, Brown Myles, Cichowski Karen, Long Henry, Bradner James E, Wong Kwok-Kin: Oncogenic Deregulation of EZH2 as an Opportunity for Targeted Therapy in Lung Cancer. Cancer discovery 6(9): 1006-21, Sep 2016.
2014
De Raedt Thomas, Beert Eline, Pasmant Eric, Luscan Armelle, Brems Hilde, Ortonne Nicolas, Helin Kristian, Hornick Jason L, Mautner Victor, Kehrer-Sawatzki Hildegard, Clapp Wade, Bradner James, Vidaud Michel, Upadhyaya Meena, Legius Eric, Cichowski Karen: PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514(7521): 247-51, Oct 2014.
Malone CF, Fromm JA, Maertens O, De Raedt T, Ingraham R, Cichowski K.: Defining key signaling nodes and therapeutic biomarkers in NF1-mutant Cancers. Cancer Discovery 4(9): 1062-73, Sep 2014.
Luscan Armelle, Shackleford Ghjuvan’ghjacumu, Masliah-Planchon Julien, Laurendeau Ingrid, Ortonne Nicolas, Varin Jennifer, Lallemand François, Leroy Karen, Dumaine Valérie, Hivelin Mikael, Borderie Didier, De Raedt Thomas, Valeyrie-Allanore Laurence, Larousserie Frédérique, Terris Benoît, Lantieri Laurent, Vidaud Michel, Vidaud Dominique, Wolkenstein Pierre, Parfait Béatrice, Bièche Ivan, Massaad Charbel, Pasmant Eric: The activation of the WNT signaling pathway is a Hallmark in neurofibromatosis type 1 tumorigenesis. Clinical cancer research : an official journal of the American Association for Cancer Research 20(2): 358-71, Jan 2014.
2013
McLaughlin Sara Koenig, Olsen Sarah Naomi, Dake Benjamin, De Raedt Thomas, Lim Elgene, Bronson Roderick Terry, Beroukhim Rameen, Polyak Kornelia, Brown Myles, Kuperwasser Charlotte, Cichowski Karen: The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer cell 24(3): 365-78, Sep 2013.
2011
Beert Eline, Brems Hilde, Daniëls Bruno, De Wever Ivo, Van Calenbergh Frank, Schoenaers Joseph, Debiec-Rychter Maria, Gevaert Olivier, De Raedt Thomas, Van Den Bruel Annick, de Ravel Thomy, Cichowski Karen, Kluwe Lan, Mautner Victor, Sciot Raf, Legius Eric: Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes, chromosomes & cancer 50(12): 1021-32, Dec 2011.
De Raedt Thomas, Walton Zandra, Yecies Jessica L, Li Danan, Chen Yimei, Malone Clare F, Maertens Ophélia, Jeong Seung Min, Bronson Roderick T, Lebleu Valerie, Kalluri Raghu, Normant Emmanuel, Haigis Marcia C, Manning Brendan D, Wong Kwok-Kin, Macleod Kay F, Cichowski Karen: Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer cell 20(3): 400-13, Sep 2011.
